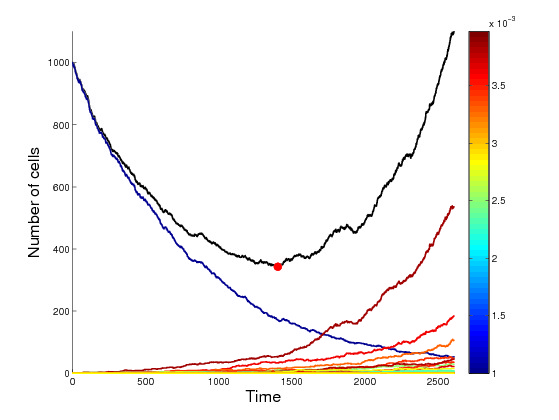
- Myklebust E, Schjesvold F, Frigessi A, Leder K, Foo J, Kohn-Luque A. Relapse prediction in multiple myeloma patients treated with isatuximab, carfilzomib, and dexamethasone, submitted (2024).
- Gunnarsson E, Kim S, Kaura K, Schmid K, Mumenthaler S, Foo J. Understanding patient-derived tumor organoid growth through an integrated imaging and mathematical modeling framework, submitted (2024).
- Wu C, Gunnarsson EB, Myklebust EM, K ̈ohn-Luque A, Tadele DS, Enserink JM, Frigessi A, Foo J, Leder K. Using birth-death processes to infer tumor subpopulation structure from live-cell imaging drug screening data. PLoS Computational Biology. 2024 Mar 6;20(3):e1011888. Link
- E. Gunnarsson, J. Foo. Mathematical models of resistance evolution under continuous and pulsed anti-cancer therapies, (invited book chapter), accepted.
- A. Li, D. Kibby, J. Foo. A Comparison of Mutation and Amplification-Driven Resistance Mechanisms and Their Impacts on Tumor Recurrence, Journal of Mathematical Biology, 87, 59 (2023). https://doi.org/10.1007/s00285-023-01992-8 Link
- Foo J, Gunnarsson E, Leder K, Sivakoff D. Dynamics of advantageous mutant spread in spatial death-birth and birth-death Moran models, Comm. Appl. Math. Comput. 2023. Link
- A. Kohn-Luque, E. M. Myklebust, D. S. Tadele, M. Gilberto, J. Noory, E. Harivel, P. Arsenteva, S. Mumenthaler, F. Schjesvold, K. Tasken, J. Enserink, K. Leder, A. Frigessi, J. Foo. Phenotypic deconvolution in heterogeneous cell populations using drug screening data, Cell Reports Methods, 2023. Link
- E. Gunnarsson, J. Foo, K. Leder. Statistical inference of the rates of cell proliferation and phenotypic switching in cancer. J. Theoretical Biology, 2023. Link
- Foo, J., Gunnarsson, E., Leder, K, and Storey, K. Spread of premalignant mutant clones and cancer initiation in multilayered tissue, Annals of Applied Probability, 2023. Link
- Foo, J., Rockne, R., Basanta, D. et. al. Roadmap on plasticity and epigenetics in cancer, Physical Biology, Vol 19 (3) 2022. Link
- Foo, J. Cancer Evolution in Spatially Structured Tissue. Notices of the AMS, 2021. Link
- Gunnarsson, E., Leder, K, and Foo. J. Exact site frequency spectra of neutrally evolving tumors, transition between power laws and signatures of cell viability. Theoretical Population Biology, Volume 142, December 2021, Pages 67-90, 2021. Link *Hon. Mention Feldman Prize
- Foo, J., Leder, K., and Schweinsberg, J. Mutation timing in a spatial model of evolution. Stochastic Processes and their Applications, Volume 130, Issue 10, Pages 6388-6413, October 2020. Link
- Kim, S., Choung, S., Sun, R., Ung, N., Hashemi, N., Lau, R., Spiller, E., Gasho, J., Foo, J., Mumenthaler, S. Comparison of cell and organoid-level analysis of patient-derived 3D organoids to evaluate tumor cell growth dynamics and drug response. SLAS Discovery, Volume: 25 issue: 7, page(s): 744-754, April 2020. Link
- Gunnarsson, E., De, S., Leder, K., and Foo, J. Understanding the role of phenotypic switching in cancer drug resistance. Journal of Theoretical Biology, Volume 490, 7 April 2020, 110162. Link
- Goldberg EE and Foo, J. Memory in trait macroevolution. American Naturalist, 2020. doi: 10.1086/705992 Link
- Storey K, Leder K, Hawkins-Daarud A, Swanson K, Ahmed A, Rockne R, Foo J. Glioblastoma recurrence and the role of MGMT methylation. JCO Clinical Cancer Informatics. 2019 (3)p. 1-12. Link
- Kaveh K, Takahashi Y, Farrar MA, Storme G, Guido M, Piepenburg J, Penning J, Foo J, Leder KZ, Hui SK. Combination therapeutics of Nilotinib and radiation in acute lymphoblastic leukemia as an effective method against drug-resistance. PLoS Computational Biology. 2017 Jul 6;13(7):e1005482. Link
- Storey K, Ryser MD, Leder K, Foo J. Spatial measures of genetic heterogeneity during carcinogenesis. Bulletin of Mathematical Biology. 2017 Feb 1;79(2):237-76. PDF
- Lindsay, D., Garvey, C., Mumenthaler, S., Foo, J. An evolutionary perspective on cancer treatment response. In: Ujvari B, Roche B, Thomas F, editors. Ecology and Evolution of Cancer. Academic Press; 2017 Feb 8.
- Lindsay, D., Foo, J. `Impact of tumor oxygenation on drug resistance evolution under hypoxia-activated prodrug therapy,' Proceedings of 5th International Conference on Biomedical Engineering, 2017.
- He, Q., Zhu, J., Dingli, D., Foo, J., Leder, K. `Optimized treatment schedules for Chronic Myeloid Leukemia,' PLoS Computational Biology (2016) Oct 20;12(10):e1005129. Link
- Ryser, M., Lee, W., Ready, N., Leder, K., Foo, J. `Quantifying the Dynamics of Field Cancerization in HPV-Ne gative Head and Neck Cancer: A Multiscale Modeling Approach,' Cancer Research (2016) doi: 10.1158/0008- 5472.CAN-16-1054. Link
- Lindsay, D., Garvey, C., Mumenthaler, S., Foo, J. `Leveraging hypoxia-activated prodrugs to prevent drug resistance in solid tumors.' PLoS Computational Biology (2016) Aug 25;12(8):e1005077. Link
- Garvey, C., Spiller, E., Lindsay, D., Chiang, C., Choi, N., Agus, D., Mallick, P., Foo, J., Mumenthaler, S.; `An image-based quantitative method for studying tumor cell dynamics driven by evolutionary selective pressures.' Scientific Reports 6 :29752 (2016). Link
- Durrett, R., Foo, J., Leder, K.; ‘Cancer initiation in spatially structured tissue,’ Journal of Mathematical Biology, (2016); Vol 72(5): pages 1369-400. PDF
- Foo, J., Liu, L., Leder, K., Riester, M., Iwasa, Y., Lengauer, C., Michor, F.; ‘An Evolutionary Approach for Identifying Driver Mutations in Colorectal Cancer,’ PLoS Computational Biology (2015) 11(9): e1004350. Link
- Mumenthaler, S., Foo, J., et. al. `The Impact of Microenvironmental Heterogeneity on the Evolution of Drug Resistance in Cancer Cells,' Cancer Informatics (2015) 4 (Suppl 4): 19–31. PDF
- Foo, J., Haskell, C., Komarova, N., Segal, R., Wood, K. `Modeling sympatric speciation in quasiperiodic environments,' IMA Volumes in Mathematics and its Applications: Applications of Dynamical Systems in Biology and Medicine, Volume 158, p. 149-174 (2015). PDF
- Foo, J., Leder, K., and Zhu, J. `Escape times for branching processes with random mutational fitness effects,' Stochastic Processes and their Applications (2014) Volume 124, Issue 11, p. 3661–3697. PDF
- Foo, J., Leder, K., and Ryser, M. `Multifocality and recurrence risk: A quantitative model of field cancerization,' Journal of Theoretical Biology (2014) Vol 355. PDF
- Aghili, L., Foo, J., De Gregori, J., and De, S. `The landscape of detectable somatically acquired amplifications and deletions in apparently normal human tissues of cancer patients,' Cell Repts (2014) Vol 7 Issue 4. PDF
- Foo, J. and Michor, F. `Evolution of acquired resistance to anti-cancer therapy,' Journal of Theoretical Biology (2014) Vol 355. link
- Foo, J. and Leder, K. `Dynamics of cancer recurrence,' Annals of Applied Probability (2013) Vol 23 (4), 1437-1468. PDF
- The Physical Sciences-Oncology Center Network (2013) A physical sciences network characterization of nonmalignant and metastatic cells. Scientific Reports 3, 1-12. Link
- Foo, J, Leder, K. and Mumenthaler, S. `Cancer as a moving target: Understanding the composition and rebound growth kinetics of recurrent tumors,' Evolutionary Applications (2013) Vol 6(1). Link
- Shu, J., Zhilian, X., Liang, E., Slipek, N,. Shen, D., Foo, J., Subramanian, S., Steer, C. `Dose-dependent differential mRNA target selection and regulation by let-7a-7f and miR-17-92 cluster microRNAs,' RNA Biology (2012) Vol 9 (10). Link
- Foo, J., Chmielecki, J., Pao, W., Michor, F. `Effects of pharmacokinetic processes and varied dosing schedules on the dynamics of acquired resistance to erlotinib in EGFR-mutant lung cancer,' Journal of Thoracic Oncology (2012) Vol 7 (10). PDF
- Tang, M., Foo, J., Gonen, M., Guilhot, J., Mahon, F., Michor, F. `Selection pressure exerted by imatinib therapy leads to disparate outcomes of imatinib discontinuation trals,' Haematalogica (2012) 2012 Vol 97(10). PDF
- Foo, J., Leder, K. `Rare events in cacner recurrence timing,'Proceediings of the 2012 Winter Simulation Conference IEEE.' (2012) 1-10.
- Mumenthaler S, Foo J, Leder K, et al. `Evolutionary modeling of combination strategies to overcome resistance to tyrosine kinase inhibitors in non-small cell lung cancer,' Mol. Pharm (2011) 8(6). Link
- Chmielecki J, Foo J, et al. `Optimization of dosing for
EGFR-mutant non-small cell lung cancer with evolutionary cancer
modeling,' Science Translational Medicine (2011) Vol 3, Issue 90:
90ra59. Link
- Leder, K., Foo, J., Skaggs, B., Gorre, M., Sawyers, C., Michor, F. `Diversity in pre-existing resistance to BCR-ABL inhibitors in Chronic Myeloid Leukemia,' PLoS ONE (2011)e27682. doi:10.1371/journal.pone.0027682. PDF SI
- Durrett, R., Foo, J. Leder, K., Mayberry, J., Michor, F. `Intra-tumor heterogeneity in evolutionary models of tumor progression' Genetics (2011) Vol 188 (2): 461-477. PDF
- Foo, J., Leder, K., and Michor, F. `Stochastic dynamics of cancer initiation,' Physical Biology (2011) Vol 8: 015002. PDF
- Foo, J. and Karniadakis, G. `Multi-Element Probabilistic Collocation in High Dimensions,' Journal of Computational Physics (2010) Vol 229(5): p.1536-1557. PDF
- Durrett, R., Foo, J. Leder, K., Mayberry, J., Michor, F. `Evolutionary dynamics of tumor progression with random fitness values' Theoretical Population Biology (2010) Vol 78: p.54-66. PDF
- Foo, J. and Michor, F. `Evolution of resistance to targeted anti-cancer therapies during general dosing schedules,' Journal of Theoretical Biology (2010) Vol 263 p. 179-188. PDF
- Foo, J. and Michor, F. `Evolution of resistance to targeted anti-cancer therapies during continuous and pulsed administration strategies,' PLoS Computational Biology (2009) 5, e1000557. Link
- Foo, J., Drummond, M., Clarkson, B., Holyoake, T., and Michor, F. `Eradication of Chronic Myeloid Leukemia Stem Cells: A Novel Mathematical Model Predicts No Therapeutic Benefit of Adding G-CSF to Imatinib,' PLoS Computational Biology (2009) 5, e10000503. PDF
- Foo, J., Sindi, S., and Karniadakis, G. `Using the Multi-Element Probabilistic Collocation Method for Sensitivity Analysis in Cellular Signaling Networks.' Systems Biology (2009) Vol 3(4):239-254. PDF
- Foo, J., Wan, X., Karniadakis, G. `A Multi-Element Probabilistic Collocation Method for PDEs with Parametric Uncertainty: Error Analysis and Applications' Journal of Computational Physics (2008) Vol 227: 9572-9595. PDF
- Prempraneerach, P., Foo, J., Triantafyllou, M., Chryssostomidis, C., Karniadakis, G. `Gradient-free Stochastic Sensitivity Analysis of the Ship-board Power System' International Simulation Multi-Conference (2008) Edinburgh, Scotland.
- Foo, J., Yosibash, Z., Karniadakis, G. `Stochastic Simulation of Riser-Sections with Uncertain Measured Pressure Loads and/or Uncertain Material Properties.' Computer Methods in Applied Mechanics and Engineering (2007) Vol 196: 4250-4271. PDF
- Bell, J., Foo, J., and Garcia, A. `Algorithm Refinement for the Stochastic Burgers Equation.' Journal of Computational Physics (2007) Vol 223 : 451-468. PDF
- Lucor, D., Foo, J., and Karniadakis, G. `Vortex mode selection of a rigid cylinder subject to VIV at low mass-damping.' Journal of Fluids and Structures (2005) Vol 20: 485-503. PDF
- Lucor, D., Foo, J., and Karniadakis, G. `Correlation Length and Force Phasing of a Rigid Cylinder Subject to VIV of Fully Coupled Fluid Structure Interactions.' in Fluid Mechanics and its Applications (2004) Vol 75 : 187-199.
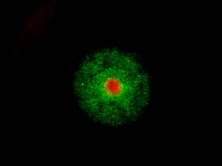
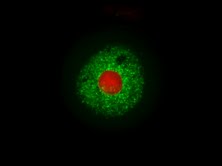 Spheroid models of resistance outgrowth.
Spheroid models of resistance outgrowth.
S. Mumenthaler (EITM)